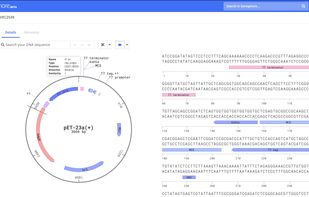
VectorBee is a highly user-friendly genetic engineering software created by VectorBuilder for viewing, editing and analyzing DNA and protein sequences.
UGENE is described as 'Is free open-source cross-platform bioinformatics software' and is an app in the education & reference category. There are more than 25 alternatives to UGENE for a variety of platforms, including Windows, Linux, Mac, Web-based and Self-Hosted apps. The best UGENE alternative is VectorBee, which is free. Other great apps like UGENE are AliView, BioEdit, Chromas and Genophore.
VectorBee is a highly user-friendly genetic engineering software created by VectorBuilder for viewing, editing and analyzing DNA and protein sequences.

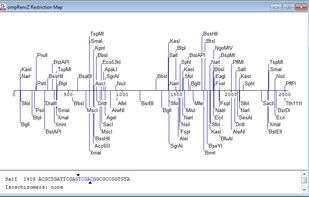
AliView is yet another alignment viewer and editor, but this is probably one of the fastest and most intuitive to use, not so bloated and hopefully to your liking.
BioEdit is a biological sequence alignment editor written for Windows 95/98/NT/2000/XP/7. An intuitive multiple document interface with convenient features makes alignment and manipulation of sequences relatively easy on your desktop computer.
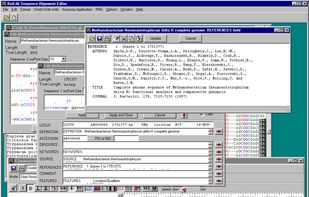
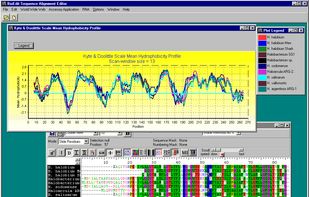
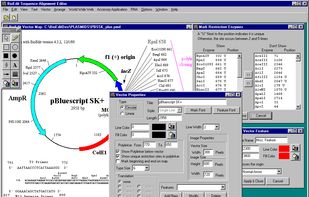
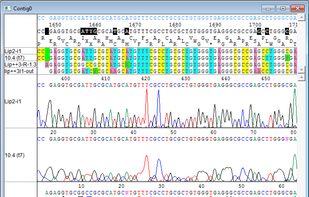
Two software applications are available from Technelysium: Chromas and ChromasPro.
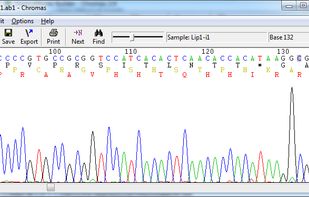
Genophore is an application for searching and designing proteins. It also provides a suite of bioinformatics utilities and collaboration tools.

RasMol is a computer program written for molecular graphics visualization intended and used primarily for the depiction and exploration of biological macromolecule structures, such as those found in the Protein Data Bank.

Geneious combines all the major DNA and protein sequence analysis tools into one revolutionary software solution!
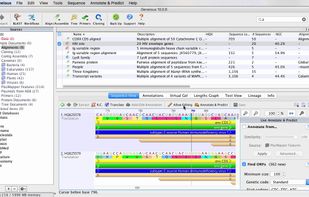
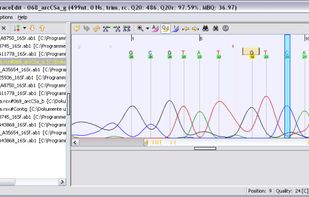
Ridom TraceEdit is a cross-platform graphical DNA trace viewer and editor. TraceEdit displays the chromatogram files from Applied Biosystems automated sequencers and files in the Staden SCF format.
A full featured multiple sequence alignment editor, analyser and shading utility for Windows.
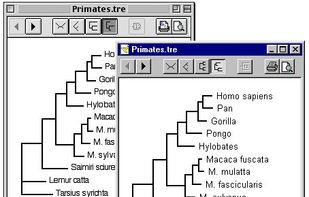
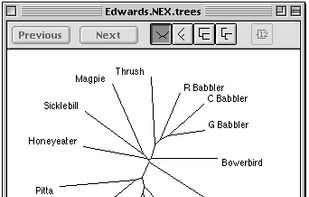
Archaeopteryx is a software tool for the visualization, analysis, and editing of potentially large and highly annotated phylogenetic trees.